Extracting structured information from clinical notes—such as identifying medical conditions, medications, and their relationships—is a cornerstone of modern healthcare research. These information extraction tasks, known as named entity recognition (NER) and relation extraction (RE), enable researchers to transform unstructured text into insights that can improve patient care and advance medical science.
Yet, developing accurate models for these tasks is difficult. Privacy concerns and the high cost of annotation mean that labeled clinical data is scarce. This shortage of training data limits the performance of traditional machine learning approaches.
To address this challenge, our engineering team at Truveta has developed a novel framework that combines the power of large language models (LLMs) with advanced deep learning techniques to enhance clinical information extraction. This work was published as a part of MedInfo 2025, “Structured LLM Augmentation for Clinical Information Extraction,” in August 2025.
Methods
We designed a two-part framework:
Data augmentation with LLMs
-
- LLMs were used to generate diverse, high-quality synthetic clinical examples.
- These examples preserved critical relationships between entities (e.g., a drug and its dosage) while introducing useful variation.
- This augmentation helped overcome the data scarcity problem without compromising contextual accuracy.
Segmentation-Based BERT for Long Documents
-
-
- Clinical notes are often lengthy, which creates challenges for standard NLP models.
- To address this, we developed a segmentation-based BERT model integrated with a BiLSTM layer, which persists global context across segments.
- Finally, specialized linear layers were applied for both NER and RE tasks.
-
This architecture allowed us to capture detailed local context while maintaining a broader view of the clinical note—essential for real-world electronic health record (EHR) applications.
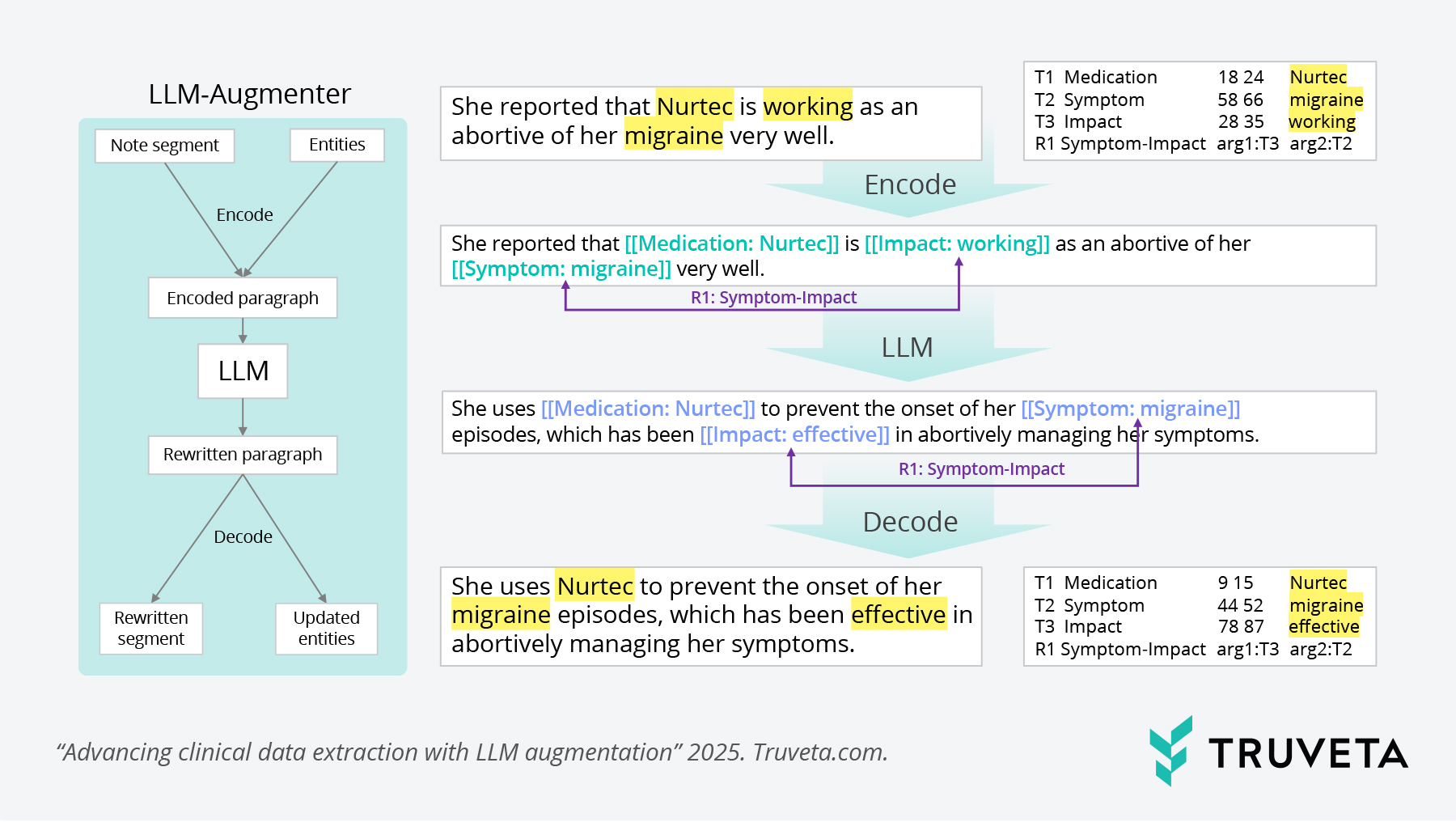
Figure 1: Illustration of augmentation process of LLM-Augmenter.
We then tested the framework across three datasets:
- i2b2-2012: 310 clinical notes with 6 entity types.
- N2C2-2018 Track 2: 505 discharge summaries with 9 entity and 8 relation types.
- Proprietary Truveta Dataset: 2,306 clinical notes with 29 entity and 17 relation types.
For each dataset, we created augmented subsets using our LLM-based process and compared results against baseline models trained only on original data.
Results
The augmented models consistently outperformed baselines across all benchmarks (with performance summarized in the table below):
- i2b2-2012: Strict NER F1 improved from 81.20% to 81.80%.
- N2C2-2018: Strict RE improved from 82.82% to 84.15%, and lenient RE rose from 90.39% to 90.76%.
- Proprietary dataset: Strict NER F1 increased from 86.22% to 87.46%, and strict RE improved from 76.68% to 77.65%.
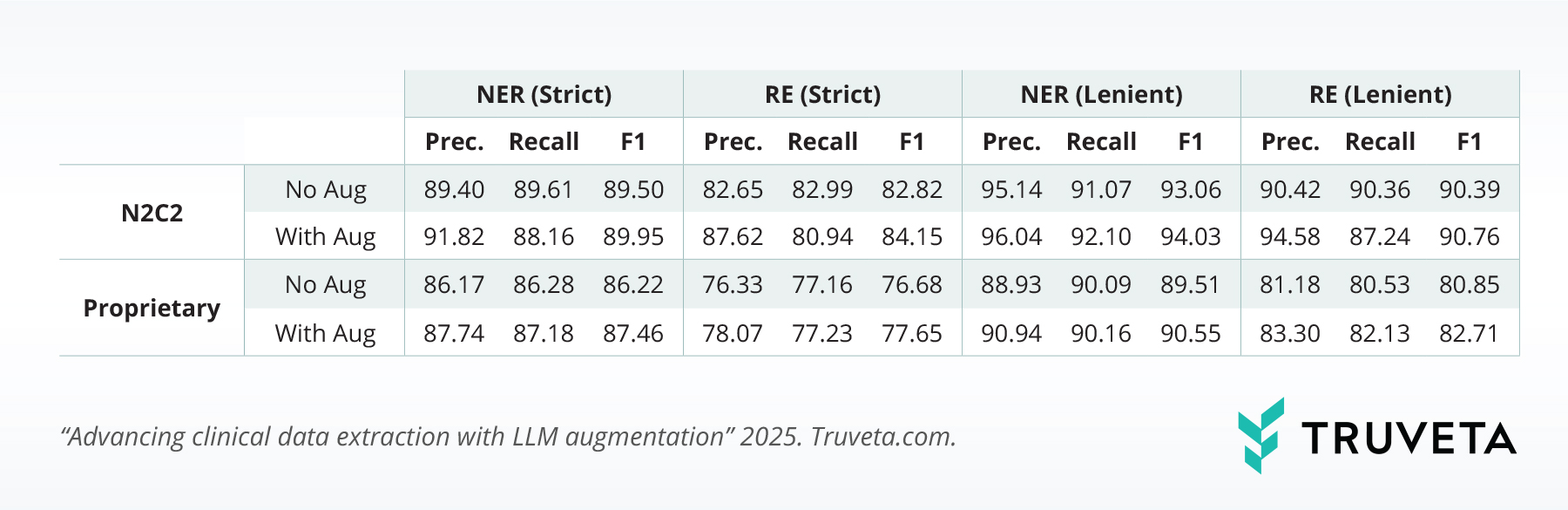
These gains—though modest in percentage terms—represent meaningful improvements in precision and recall, especially for low-resource datasets.
Discussion
By generating diverse but consistent training examples, our approach improved model generalizability across multiple benchmarks. Importantly, the segmentation-based BERT with BiLSTM proved effective at handling long clinical documents, making the framework scalable to real-world EHR datasets.
That said, we did observe a slight trade-off: in strict evaluation settings, recall occasionally dipped, reflecting the tension between diversity and sensitivity. Future work will explore strategies to fine-tune this balance.
Conclusion
Our study demonstrates that combining LLM-based data augmentation with a segmentation-based BERT architecture significantly enhances performance on key clinical information extraction tasks.
This work not only addresses two critical barriers—data scarcity and long-document complexity—but also lays the foundation for more robust, scalable, and clinically useful natural language processing systems.
At Truveta, we believe these advancements bring us one step closer to unlocking the full potential of clinical data to improve patient outcomes.
References
[1] N. Perera, M. Dehmer, and F. Emmert-Streib. Named entity recognition and relation detection for biomedical information extraction. Frontiers in Cell and Developmental Biology, 2020.
[2] Z. Meng, T. Liu, H. Zhang, K. Feng, and P. Zhao. CEAN: Contrastive Event Aggregation Network with LLM-based Augmentation for Event Extraction. Proceedings of the 18th Conference of the European Chapter of the Association for Computational Linguistics, 2024.
[3] M. Zhang, G. Jiang, S. Liu, J. Chen, and M. Zhang. LLM–assisted data augmentation for Chinese dialogue–level dependency parsing. Computational Linguistics, 2024.
[4] J. Dagdelen, A. Dunn, S. Lee, N. Walker, A. S. Rosen, G. Ceder, K. A. Persson, and A. Jain. Structured information extraction from scientific text with large language models. Nature Communications, 2024.
[5] J. Devlin. BERT: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint, 2018.
[6] I. Beltagy, M. E. Peters, and A. Cohan. Longformer: The long-document transformer. arXiv preprint arXiv:2004.05150, 2020.
[7] Y. Peng, S. Yan, and Z. Lu. Transfer Learning in Biomedical Natural Language Processing: An Evaluation of BERT and ELMo on Ten Benchmarking Datasets. 2019.
[8] A. E. Johnson, T. J. Pollard, L. Shen, L.-w. Lehman, M. Feng, M. Ghassemi, B. Moody, P. Szolovits, L. A. Celi, and R. G. Mark. MIMIC-III, a freely accessible critical care database. Scientific Data, 2016.
[9] S. Siami-Namini, N. Tavakoli, and A. S. Namin. The performance of LSTM and BiLSTM in forecasting time series. 2019 IEEE International Conference on Big Data (Big Data), 2019.
[10] S. Henry, K. Buchan, M. Filannino, A. Stubbs, and O. Uzuner. 2018 n2c2 shared task on adverse drug events and medication extraction in electronic health records. Journal of the American Medical Informatics Association, 2020.
[11] S. Henry, K. Buchan, M. Filannino, A. Stubbs, and O. Uzuner. 2018 n2c2 shared task on adverse drug events and medication extraction in electronic health records. Journal of the American Medical Informatics Association, 2020.

