Authors: Wilson Lau, PhD ⊕Truveta, Inc, Bellevue, WA,, Youngwon Kim, PhD ⊕Truveta, Inc, Bellevue, WA, Sravanthi Paras, MD ⊕Swedish Medical Center, Seattle, WA, Md Enamul Haque, PhD ⊕Truveta, Inc, Bellevue, WA,, Sara Daraei, PhD ⊕Truveta, Inc, Bellevue, WA, Rajesh Rao, MS ⊕Truveta, Inc, Bellevue, WA,, Jay Pillai ⊕Truveta, Inc, Bellevue, WA, Anand Oka, PhD ⊕Truveta, Inc, Bellevue, WA
- Early-onset colorectal cancer (EoCRC) is rising in adults under 50, yet current screening guidelines typically exclude this age group.
- This study investigated the potential of machine learning (ML) and large language models (LLMs) to predict the risk of early-onset colorectal cancer (EoCRC) in individuals aged 18–44, utilizing the electronic health record (EHR) data within the last six months of the patient journey prior to CRC diagnoses.
- A fine-tuned GPT-4o LLM outperformed other models—achieving 90%+ specificity even when disease prevalence was just 1%—and could help reduce unnecessary screenings.
- These findings suggest LLMs may be a powerful tool for identifying rare cancers earlier in younger patients not covered by current screening protocols.
CRC poses a significant public health challenge in the United States, being the second leading cause of cancer-related mortality and the fourth most common new cancer diagnosis in 2024 (Siegel et al., 2024). While being traditionally a disease of older adults, an alarming trend showed increasing incidence of CRC in individuals under 50. Annually, CRC incidence in this age group increased by 1.9% between 2011 and 2019, while advanced-stage CRC among those aged 20-49 saw an approximate 3% yearly rise from 2012 to 2021 (Siegel et al., 2023).
Given that current screening guidelines recommend initiation at age 45, younger individuals tend to be diagnosed at more advanced and less treatable stages (di Martino, 2022). These concerning trends highlight a critical need for additional strategies for early identification in younger populations to facilitate timely intervention and enhance outcomes (Townsend et al., 2021; Meystre et al., 2017). By integrating diverse clinical and demographic data, EHR-based predictive approaches have the potential to enhance early detection, and facilitate timely medical evaluations, thereby mitigating the impact of EoCRC.
ML methods are highly capable of modeling complex, non-linear relationships and interactions within large datasets (Zhen et al., 2024). The recent advancement in LLMs has further expanded these predictive capabilities in clinical research (Qiu, 2023). LLMs have yielded promising results in the prediction of diverse health conditions—including depression, sleep disorders, and stress levels—and in the diagnosis of rare diseases, demonstrating their potentials to identify subtle patterns within clinical data.
Motivated by these advancements, our study utilized both ML and LLM approaches to investigate their capability in EoCRC risk prediction. This work is in collaboration with Sravanthi Parasa, MD — a specialist in Gastroenterology from Swedish Medical Center.
Our research is guided by the following questions:
Research questions
- Can EHR data be effectively utilized to predict CRC risk in a younger population (ages 18-44)?
- How do statistical ML and LLM compare in their ability to predict CRC risk in individuals aged 18-44 using EHR data?
- Given that early-onset CRC is typically a rare condition in real-world settings, to what extent does the variation in CRC prevalence influence the predictive performance metrics (e.g., precision, recall, F1-score, specificity) of these different modeling approaches?
Methods
These studies were conducted to evaluate and compare the performance of ML and LLM in predicting EoCRC risk using EHR data. We designed two distinct studies with varying CRC prevalence: a non-rare CRC population and a rare CRC population.
Study 1: Non-Rare CRC (Higher Prevalence)
- Training: 1,097 CRC samples (25%), 3,350 Non-CRC samples (75%).
- Test: 279 CRC samples (20%), 826 Non-CRC samples (80%).
- Features: Medical conditions, laboratory results.
Study 2: Rare CRC (Lower Prevalence)
- Training (Balanced): 1,853 CRC samples (50%), 1,835 Non-CRC samples (50%).
- Test (10 sets): 10 CRC samples (1%), 990 Non-CRC samples (99%) per set
- Features: Medical conditions, laboratory results, clinical observations.
Patient selection for both studies followed consistent criteria: CRC cohorts included individuals aged 18-44 years at confirmed diagnosis, except those with personal or family CRC history, Crohn’s disease, Lynch syndrome, or ulcerative colitis. Non-CRC controls were drawn from the same age group and did not have these conditions. To analyze these datasets, multiple predictive ML algorithms were experimented, including Random Forest and XGBoost, as well as OpenAI’s LLM GPT-4o (version 2024-08-06). A Chain-of-Thought prompting strategy, incorporating detailed instructions and CRC-specific knowledge, was employed to guide the LLM.
Results
Study 1: Non-rare CRC scenario (higher prevalence)
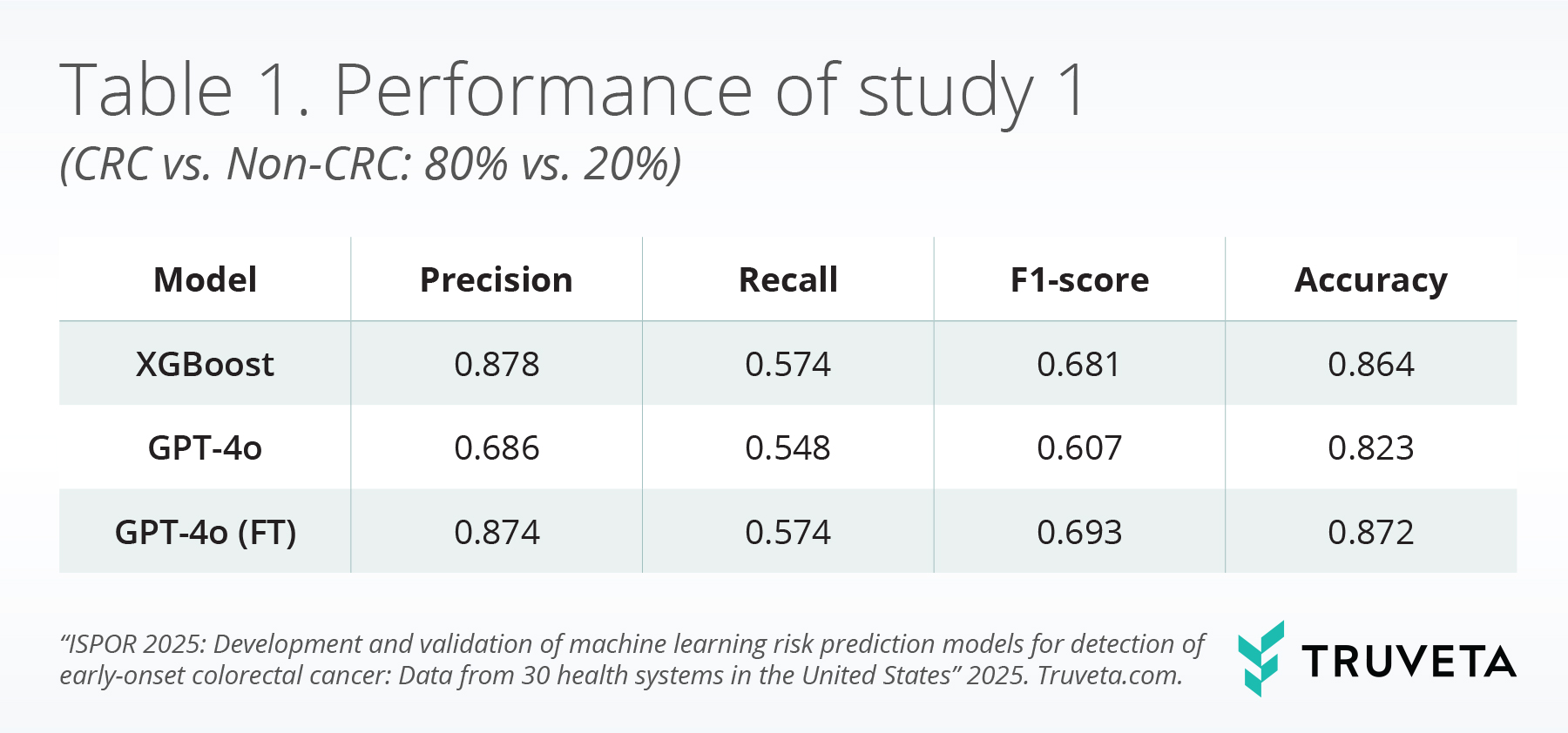
In the scenario with a higher prevalence of CRC (approximately 20% in the test set), both the XGBoost model and the fine-tuned GPT-4o model demonstrated comparable performances, outperforming the base GPT-4o model. As shown in Table 1, the fine-tuned GPT-4o achieved the highest F1-score (0.693) and overall accuracy (87.2%). This indicates that the fine-tuned model can identify over half of the EoCRC patients with a relatively low number of false positives.
Study 2: Rare CRC scenario (lower prevalence)
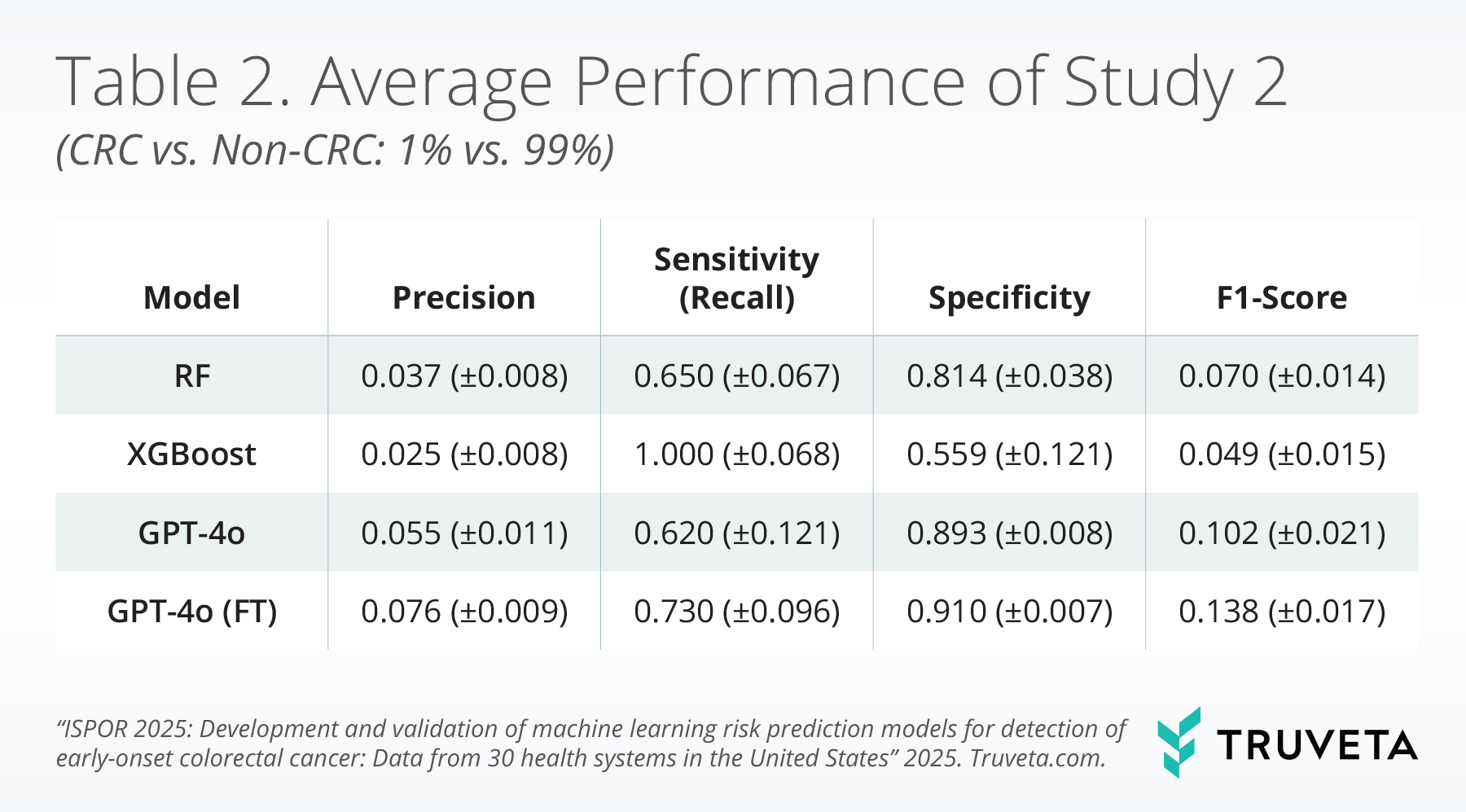
Table 2 presents the average model performance across 10 test runs in the rare disease scenario (1% CRC prevalence). Given the significant class imbalance, all models, as expected, exhibited limited precision and F1-scores. However, key differences emerged in their balance of sensitivity (recall) and specificity.
The fine-tuned GPT-4o model demonstrated the best overall performance, achieving the highest specificity (0.910), alongside a strong sensitivity of 0.730. Supervised fine-tuning notably improved the base LLM’s performance. The performance of base GPT-4o was generally comparable to or slightly better than Random Forest. While the XGBoost model achieved perfect sensitivity (1.000), this came at the cost of lower specificity (0.559). Ultimately, the fine-tuned GPT-4o offered the most reliable trade-off in identifying CRC and non-CRC samples, suggesting its potential utility in real-world screening for rare conditions.
Discussion
Our investigation into the application of ML and LLMs for EoCRC risk prediction using EHR has yielded several important insights. We believe this work underscores the significant potential of applying these advanced analytical techniques in addressing the concerning rise of EoCRC in younger population, who are not covered by current screening guidelines.
A key finding is the promising performance of a fine-tuned LLM in comparison to established ML models like XGBoost and Random Forest in predicting EoCRC across the different scenarios. Interestingly, the base GPT-4o model demonstrated comparable performance to the Random Forest model in the rare CRC study, even without specific model training. This suggests the potential utility of LLMs in rare diseases with limited data availability.
Beyond predictive accuracy, LLMs offer an advantage in interpretability. While statistical tree-based ML algorithms offer analytical interpretation, such as feature importance for global understanding and SHAP values for individual prediction explanation, LLMs, in contrast, can generate more intuitive, summary-based explanations for their predictions.
The full study is currently under peer review, and the complete paper will be shared upon its acceptance or publication.
Citations
Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA: a cancer journal for clinicians. 2024;74(1):12-49.
Siegel RL, Wagle NS, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2023. CA: a cancer journal for clinicians. 2023;73(3):233-54.
di Martino E, Smith L, Bradley SH, Hemphill S, Wright J, Renzi C, et al. Incidence trends for twelve cancers in younger adults—a rapid review. British journal of cancer. 2022;126(10):1374-86.
Townsend JS, Jones MC, Jones MN, Waits AW, Konrad K, McCoy NM. A case study of early-onset colorectal cancer: using electronic health records to support public health surveillance on an emerging cancer control topic. Journal of registry management. 2021;48(1):4.
Meystre SM, Lovis C, B¨urkle T, Tognola G, Budrionis A, Lehmann CU. Clinical data reuse or secondary use: Current status and potential future progress. Yearbook of medical informatics. 2017;26(01):38-52.
Zhen J, Li J, Liao F, Zhang J, Liu C, Xie H, et al. Development and validation of machine learning models for young-onset colorectal cancer risk stratification
Qiu J, Li L, Sun J, Peng J, Shi P, Zhang R, et al. Large ai models in health informatics: Applications, challenges, and the future. IEEE Journal of Biomedical and Health Informatics. 2023;27(12):6074-87.


